DOP30 Multi-‘omics profiling in patients with quiescent inflammatory bowel disease identifies biomarkers predicting relapse
N. Borren1, D. Plichta2, A. joshi1, G. Bonilla3, R. Sadreyev3, H. Vlamakis2, R. Xavier1, A. Ananthakrishnan1
1Massachusetts General Hospital, Division of Gastroenterology, Boston, USA, 2Infectious Disease and Microbiome Program, Broad Institute of MIT and Harvard, Cambridge, USA, 3Department of Molecular Biology, Massachusetts General Hospital, Boston, USA
Background
Inflammatory bowel diseases (IBD) are characterised by intermittent relapses and their course is heterogeneous and often unpredictable. The ability of protein, metabolite, or microbial biomarkers to predict relapse in patients with quiescent disease is unknown.
Methods
This prospective study enrolled patients with quiescent Crohn’s disease (CD) and Ulcerative colitis (UC) defined as absence of clinical symptoms (HBI < 4, SCCAI < 2) and a colonoscopy within the 1 year prior with endoscopic remission. The primary outcome was relapse within 2 years, defined as symptomatic worsening accompanied by elevated inflammatory markers resulting in a change in therapy, or IBD-related hospitalisation or surgery. Metabolomic and proteomic profiling was performed on serum samples and stool samples underwent shotgun metagenomic sequencing on Illumina HiSeq platform. Biomarkers were tested in a derivation cohort and their performance examined in an independent validation cohort.
Results
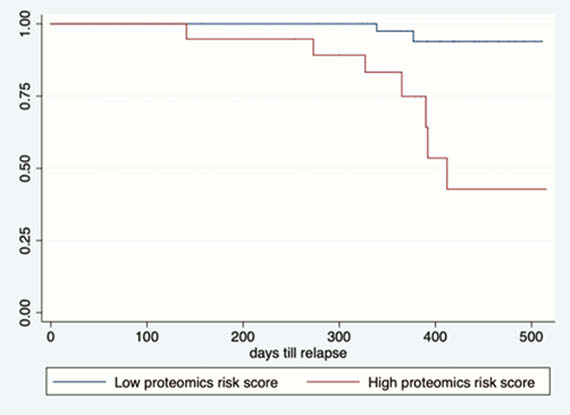
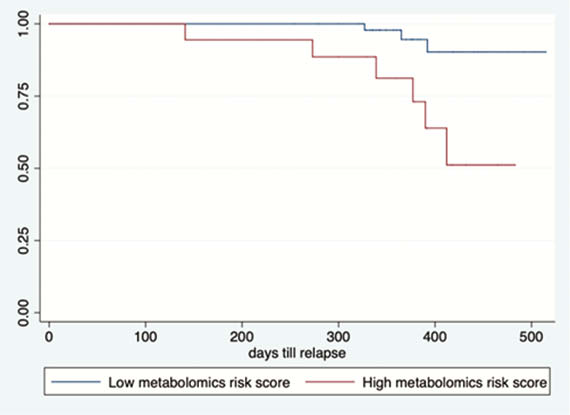
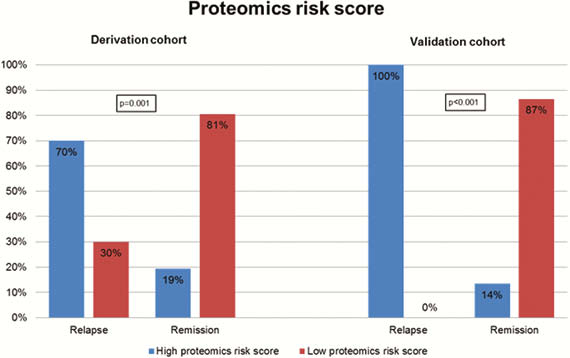
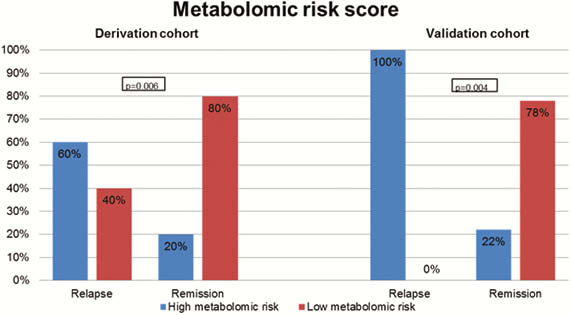
Our prospective cohort study included 164 patients with IBD (108 CD, 56 UC). Upon follow-up for a median of 1 year, 22 patients (13.4%) experienced a relapse. Three protein biomarkers (IL-10, GDNF, and CD8A) and four metabolomic markers (propionyl-
Conclusion
Proteomic, metabolomic, and microbial biomarkers identify a pro-inflammatory state in quiescent IBD that predisposes to clinical relapse.


