DOP46 Impact of a genetic burden on familial aggregation of inflammatory bowel disease
H.S. Lee1,2, L. Hannes1, M. Vancamelbeke3, V. Ballet4, M. Ferrante3,4, S. Vermeire3,4, I. Cleynen1
1Laboratory of Complex Genetics, Department of Human Genetics, KU Leuven, Leuven, Belgium, 2Department of Biochemistry and Molecular Biology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea Republic of, 3Department Chronic Diseases Metabolism and Ageing CHROMETA, Translational Research Center for Gastrointestinal Disorders TARGID, KU Leuven, Leuven, Belgium, 4Department of Gastroenterology and Hepatology, University Hospitals Leuven, Leuven, Belgium
Background
Family history of inflammatory bowel disease (IBD) is the strongest risk factor for IBD. There currently, however, is limited understanding of the contribution of genetic risk to familial aggregation of IBD. We aimed to evaluate the association between the IBD polygenic risk score (PRS) and familial IBD, and determine its contribution to familial IBD.
Methods
We included 54 multiple-affected families (≥3 first-degree relatives affected) of European ancestry, including 189 affected IBD patients (156 Crohn’s disease; 33 ulcerative colitis), and 133 unaffected relatives. For all individuals, Immunochip genotypes were available. Weighted PRSs with estimates derived from literature were calculated using PRSice-2.0, including clumping and different
Results
Using pT = 0.05 for PRS calculation, we found that affected relatives had a higher PRS than unaffected relatives (
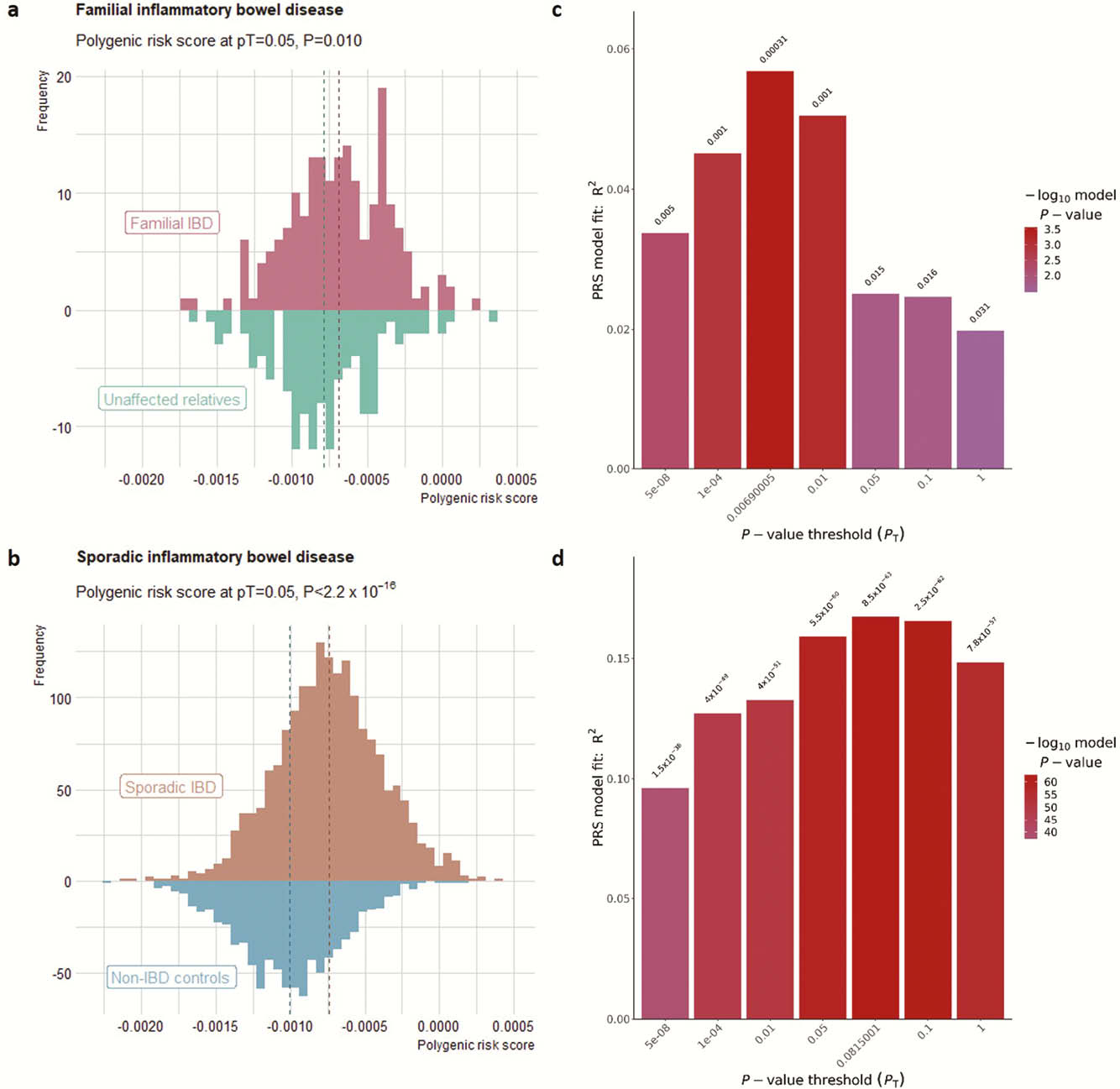
Conclusion
Higher IBD polygenic risk increases the risk for familial IBD as it does for sporadic IBD. In sporadic IBD, the increased risk is defined by variants of all


