OP29 Peripheral blood DNA methylation biomarkers accurately predict clinical- and endoscopic response to vedolizumab in a real-life cohort of Crohn’s Disease patients
Joustra, V.(1);Hageman, I.(1,2);Li Yim, A.(2,3);Levin, E.(4,5);Satsangi, J.(6);Adams, A.(6);De Jonge, W.(2);Henneman, P.(3);D'Haens, G.(1);
(1)Amsterdam UMC- University of Amsterdam, Department of Gastroenterology & Hepatology, Amsterdam, The Netherlands;(2)Amsterdam UMC- University of Amsterdam, Tytgat Institute for Liver and Intestinal Research, Amsterdam, The Netherlands;(3)Amsterdam UMC- University of Amsterdam, Genome Diagnostics Laboratory- Department of Clinical Genetics, Amsterdam, The Netherlands;(4)Horaizon, Bv, Delft, The Netherlands;(5)Amsterdam UMC- University of Amsterdam, Department of Vascular Medicine, Amsterdam, The Netherlands;(6)Oxford University- Hospitals NHS Foundation Trust- John Radcliffe Hospital, Translational Gastroenterology Unit- NIHR Oxford Biomedical Research Centre, Oxford, United Kingdom; on behalf of the EPIC consortium
Background
Despite the proven efficacy of vedolizumab (VDZ), only 29% and 36% of the Crohn’s disease (CD) patients present corticosteroid-free clinical- and endoscopic remission, respectively. Therefore, predictive biomarkers for treatment success would be of extreme value. Previous studies have identified aberrant DNA methylation associated with CD-specific phenotypes, suggesting that the methylome may be useful for classification and prediction of VDZ treatment response. Here, we sought to identify such DNA methylation biomarkers that can predict clinical- and endoscopic response to VDZ in CD patients.
Methods
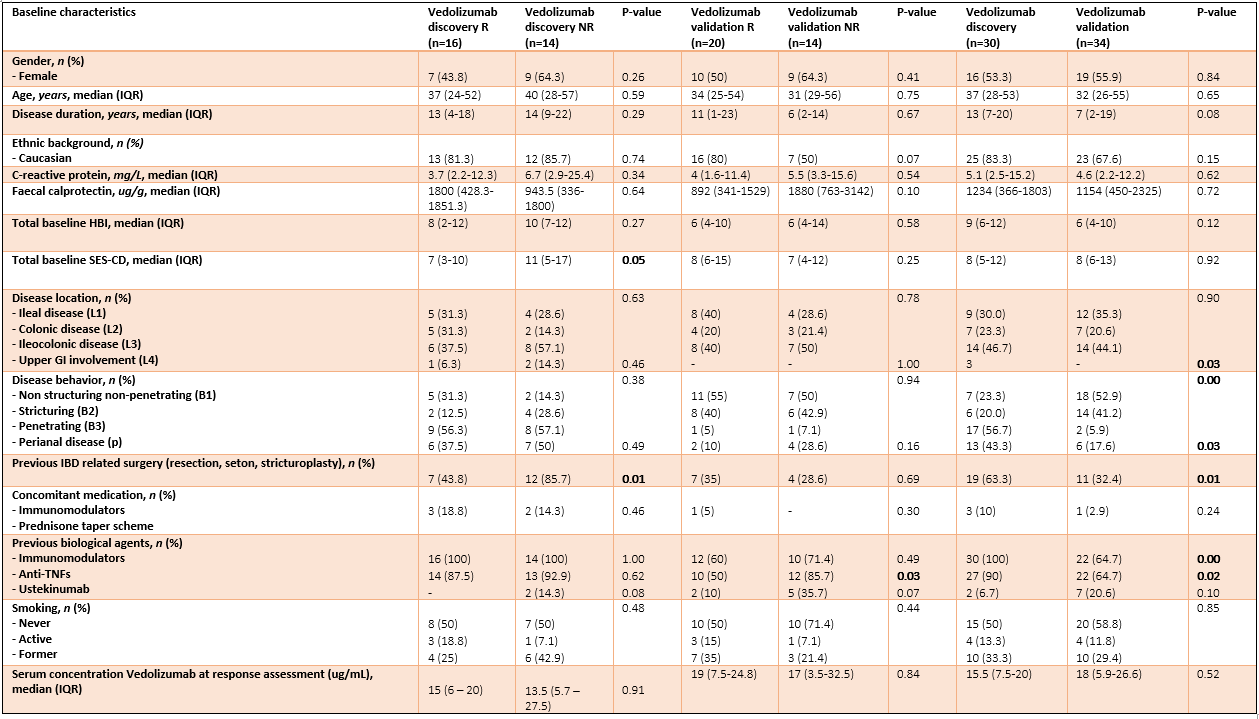
We prospectively recruited adult CD patients that initiated VDZ treatment following a baseline colonoscopy in two cohorts: a discovery and validation cohort. Peripheral blood DNA methylation profiles were measured prior to treatment (T1), and after a median of 22 weeks (T2) using the Illumina Infinium HumanMethylation EPIC BeadChip array. Response (R) was defined as the strict combination of endoscopic- (≥50% reduction in SES-CD score) and steroid-free clinical response (≥3 point drop in HBI and HBI ≤4 AND no systemic steroids) and/or biochemical response (≥50% reduction in C-reactive protein (CRP) and fecal calprotectin or a CRP ≤5 g/mL and fecal calprotectin ≤250 µg/g). Twenty-one patients had deep remission (DR), defined as a combined endoscopic- (SES-CD≤2) and steroid-free clinical remission (HBI ≤4, no systemic steroids). Biomarker identification and classification analyses were performed using stability selection gradient boosting.
Results
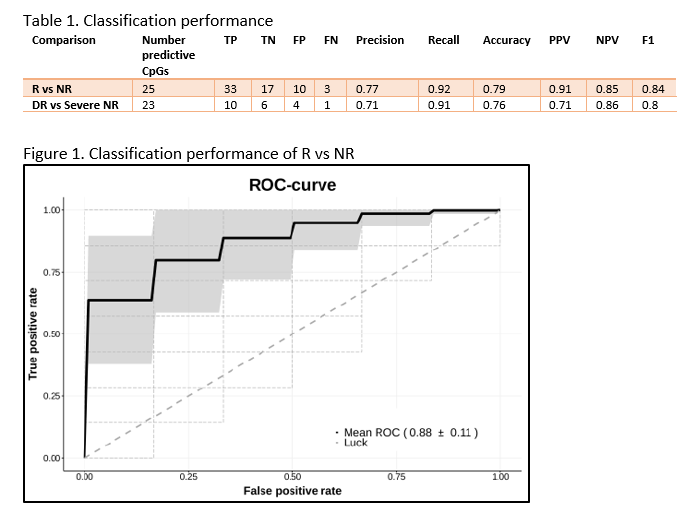
In total, 64 CD patients were enrolled (discovery 16R/14NR and validation 20R/14NR). Both cohorts were comparable for age, sex and smoking status. Forty-nine (77%) patients had previously failed an anti-TNF agent. All patients had measurable serum vedolizumab concentration at T2 (median 14.5 (6.9 – 21.3) µg/mL). Through classification analysis at T1, we were capable of discriminating R from NR with high predictive performance (25 CpGs, AUC 0.88, F1-score of 0.84, PPV of 0.91 and NPV of 0.85). When analysing the methylome of patients in deep remission, we identified 23 CpGs with high predictive performance upon independent validation (F1-score 0.80, PPV of 0.71 and NPV of 0.86). Investigating the CpGs of interest implicated genes involved in endothelial cell-cell adhesion and integrin dependent T-cell homing, corroborating VDZ’s mode of action.
Conclusion
We demonstrate two novel 25- and 23-feature panels of epigenetic biomarkers that accurately predict response or deep remission to vedolizumab respectively. Similar analyses on infliximab, adalimumab and ustekinumab are currently ongoing as part of the EPIC-CD study.