P004 Microbiota, not host origin drives ex vivo epithelial response in ulcerative colitis patients and non-IBD controls.
Arnauts, K.(1);Sudhakar, P.(1);Verstockt, S.(1);Lapierre, C.(1);Potche, S.(1);Caenepeel, C.(1);Verstockt, B.(1,2);Raes, J.(3);Vermeire, S.(1,2);Sabino, J.(1,2);Verfaillie, C.(4);Ferrante, M.(1,2);
(1)KU Leuven, TARGID- Department of Chronic Diseases- Metabolism & Ageing, Leuven, Belgium;(2)UZ Leuven, Department of Gastroenterology and Hepatology- University Hospitals Leuven, Leuven, Belgium;(3)KU Leuven, Department of Microbiology and Immunology- Rega Institute, Leuven, Belgium;(4)KU Leuven, Stem Cell Institute Leuven- Department of Development and Regeneration- Leuven- Belgium., Leuven, Belgium;
Background
Host-microbial interactions in inflammatory bowel disease (IBD) are poorly understood. Furthermore, patients with IBD have microbial dysbiosis and their epithelial cells exhibit intrinsic defects. We aimed to unravel the effect of exposure to microbiota on epithelial cells from both patients with ulcerative colitis (UC) and non-IBD controls.
Methods
Confluent Transwell® organoid-derived monolayers of 8 UC patients and 8 non-IBD controls were co-cultured for 6 hours with microbiota (3.108 cells) derived from active UC patients (n=3, endoscopic Mayo score ≥2) or healthy volunteer (HV, n=1, selected on high microbial cell count and presence of selected phyla1). For this purpose, fresh faecal samples were filtered and frozen in 0.9% NaCl. Inflammation was re-induced with 100 ng/ml TNF-α, 20 ng/ml IL-1β, and 1 µg/ml flagellin in confluent Transwell® cultures, 24 hours prior to microbiota co-culture. Transepithelial electrical resistance (TEER), 4 kDa fluorescein isothiocyanate (FITC) dextran (2 mg/ml) measurements, and RNA sequencing by Truseq were performed on epithelial cells, and 16S rRNA sequencing on microbiota samples before and after co-culture.
Results
The transcriptomic response of epithelial cells to microbiota was clearly influenced by the type of microbiota stimulation (no, UC or HV microbiota). However, this response was not influenced by the origin of the epithelial cells (UC or non-IBD controls) (Figure 1A). TEER and FITC dextran measurements showed a strong decrease in epithelial integrity (in both UC and non-IBD epithelium) following stimulation with UC microbiota, in contrast to HV microbiota (Figure 1B). In UC epithelial cells, stimulation with microbiota from UC patients induced a stress induced phenotype including activation of EGR1 signalling, AP-1 family, FOSL genes, MAPK and JNK signalling (Figure 2). Activation of stress pathways (MAPK family cascades) and key markers (e.g. FOSB, FOSL1, MYC, EGR1, ATF3, NOTCH1) was lower after HV microbiota stimulation (Figure 3-4). Expression levels of key markers were UC microbiota specific and similar in epithelial cells of UC and non-IBD controls (Figure 4).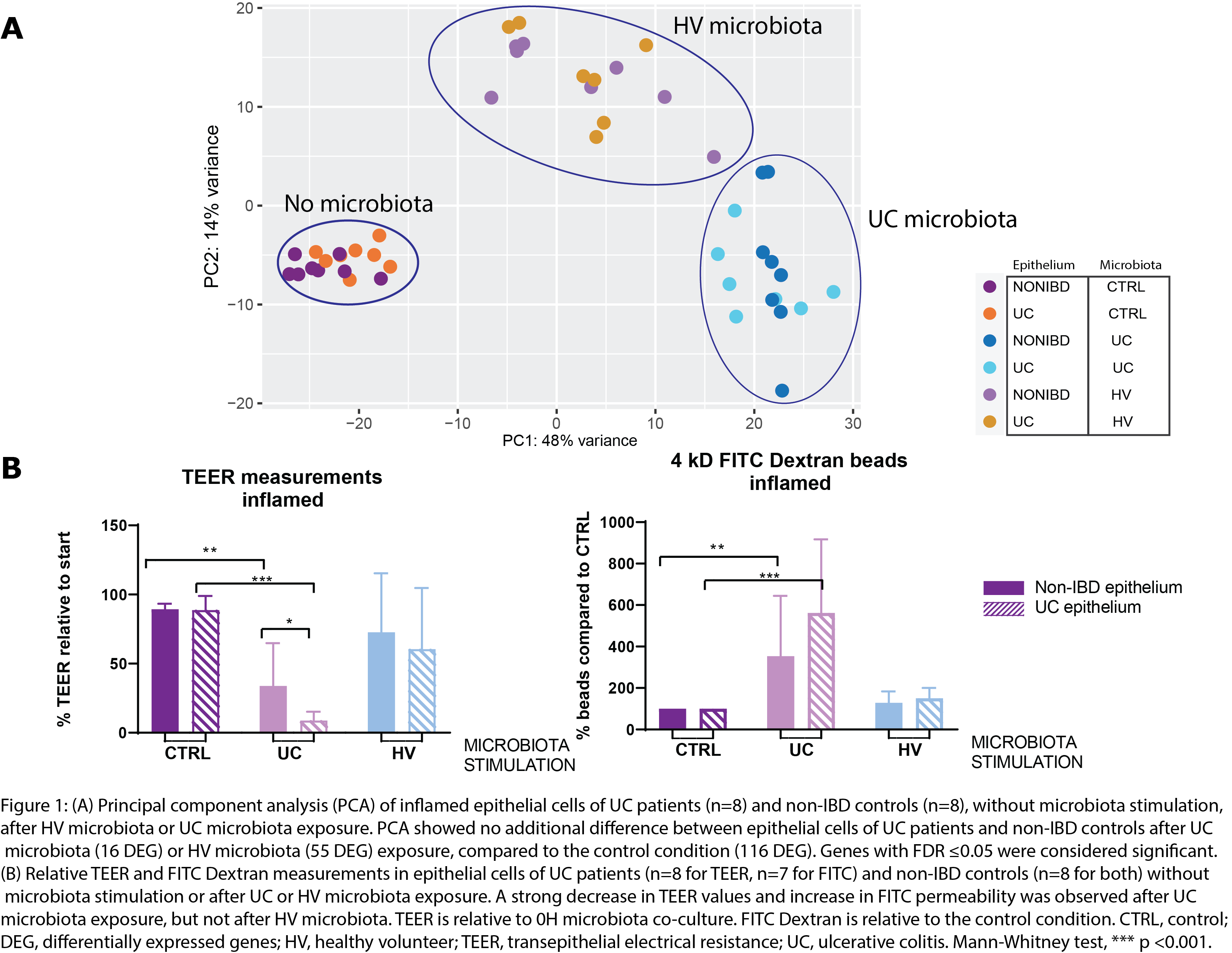



Conclusion
Not the host (UC vs non-IBD epithelial cells) but the microbial donor (UC vs HV) is driving the transcriptomic response. Epithelial cells from UC patients did not show an increased sensitivity towards microbiota stimulation compared to non-IBD epithelial cells. In contrast, exposure of epithelial cells to UC microbiota was sufficient to induce a strong stress response and barrier disruption. Further research on therapies to restore the microbial balance, to remove the constant trigger of dysbiosed microbiota, is required.
1.Vermeire, JCC, 2015


