P024 CD40 and key CD40/CD154 pathway members are significantly upregulated in inflamed lesional IBD biopsies as shown by whole transcriptome AmpliSeq gene expression profiling and corresponding immunohistochemistry.
Raulf, F.(1);Delucis-Bronn, C.(1);Angevaare, E.(1);Caesar, N.(1);Marcantonio, A.(1);Ceci, M.(1);Regnier, C.(1);Bonal, C.(2);Siegel, R.(2);Niess, J.H.(3);Wieczorek, G.(1);Hrúz, P.(3);
(1)Novartis Pharma AG / Novartis Institutes for BioMedical Research, Autoimmunity- Transplantation and Inflammation Disease Area, Basel, Switzerland;(2)Novartis Pharma AG / Novartis Institutes for BioMedical Research, Translational Medicine, Basel, Switzerland;(3)Clarunis AG, University Center for Gastrointestinal and Liver Diseases, Basel, Switzerland;
Background
Inflammatory bowel diseases (IBDs) are chronic inflammatory disorders with growing evidence of a critical role of the CD40/CD40L pathway (Shenhaji et al Front Immunol 6:529). To confirm and extend these findings, we profiled colonic biopsies from IBD patients with active ulcerative colitis (UC) and Crohn’s disease (CD) as well as non-IBD controls (HV).
Methods
Mucosal inflamed and non-inflamed biopsies of IBD patients and of healthy control subjects were immediately immersed in RNAlater (for RNA) or formaldehyde (for histology). RNA was extracted by RNeasy (Qiagen) with DNase digestion, quantified by UV spectrophotometry, and quality-controlled by Bioanalyzer (Agilent). 10 ng total RNA was subjected to ultrahigh-multiplexed RT-PCR using the AmpliSeq Transcriptome Human Gene Expression kit for 20,802 genes (Thermo Fisher). Barcoded samples were multiplexed and amplicons sequenced on an Ion GeneStudio S5 (Thermo Fisher). The resulting Excel matrix of normalized reads [RPM = Reads Per Million mapped reads; equivalent to TPM, Transcripts Per Million] was analyzed by Qlucore Omics Explorer 3.7 (Qlucore) and GraphPad Prism 9 (GraphPad Software). For histology, 2 μm sections of FFPE colonic biopsies were subjected to standard hematoxylin and eosin stain, and to automated immunohistochemistry (IHC; Ventana Discovery XT & Leica BOND RX) using standard cell-type specific antibodies and anti-CD40 Ab213205 (Abcam) and anti-CD40L AF617 (R&D Systems).
Results
161 colonic biopsies were analyzed from 36 CD and 26 UC patients (from inflamed and non-inflamed mucosa per subject), and 14 HV. Unbiased gene expression analysis by principal component analysis and hierarchical clustering showed upregulation of known inflammation markers and inflammasome components. In inflamed mucosa mRNA levels of CD40 (figure), CD154, CD44, CD86, KYNU, FCRL4, and others revealed a significant consistent upregulation of the CD40/CD154 pathway. Representative corresponding IHC stainings confirmed the upregulation of CD40 and CD154 at the protein level showing strong expression of CD40 in lesional biopsies (inflamed areas and epithelial cells), moderate expression in non-lesional and low in HV biopsies (no expression in epithelial cells), while higher expression of CD154 was observed in lesional and non-lesional biopsies compared to HV. 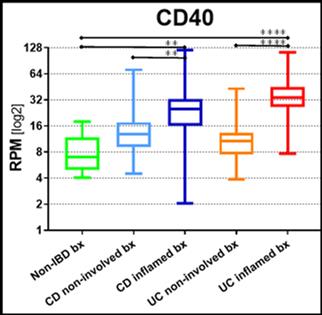
Conclusion
Consistent upregulation and activation of the CD40/CD154 axis was observed in inflamed mucosa of patients with IBD. Together with other (pre-)clinical evidence pointing to the importance of CD40 signaling, manipulation of this pathway might be a potential therapeutic target in IBD.


