P040 Proteomic characterisation of serum extracellular vesicles in patients with inflammatory bowel diseases: a novel approach for biomarker discovery
M. Chaparro1, M. Azkargorta2, A. Lapitz3, L. Ortega-Moreno1, I. Iloro2, A. Arbelaiz3, I. Escobes2, S. Fernández-Tomé1, A.C. Marin1, M. Baldan-Martin1, J.M. Falcon-Perez2, L. Bujanda3, D. Bernardo1, J. Banales3, F. Elortza2, J.P. Gisbert1
1Hospital Universitario de La Princesa, Instituto de Investigación Sanitaria Princesa IIS-IP, Universidad Autónoma de Madrid, and Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas CIBERehd, Gastroenterology, Madrid, Spain, 2CIC bioGUNE, CIBERehd, ProteoRed-ISCIII, Proteomics Platform, Derio, Spain, 3Biodonostia Research Institute. Donostia University Hospital and CIBERehd, Liver Diseases Group, San Sebastian, Spain
Background
Our aim was to identify novel biomarkers and molecular pathways that may be involved in the development or progression of inflammatory bowel diseases (IBD) by the comparison of serum extracellular vesicles (EVs) proteomic profile between healthy controls (HC), Crohn’s disease (CD) [active (aCD) and quiescent (qCD)] and ulcerative colitis (UC) [active (aUC) and quiescent (qUC)] patients.
Methods
A total of 150 individuals with an ileocolonoscopy performed the same day of blood collection were included: 30 HC, 30 aCD, 30 qCD, 30 aUC and 30 qUC. Severity of inflammation was rated based on the Simple Endoscopic Score (SES-CD) in CD and the endoscopic subscore of the Mayo index in UC. Serum EVs were isolated from blood serum and their size, distribution and concentration were determined by nanoparticle tracking analysis with NanoSight LM10 (Malvern, UK). The presence of EVs positive and negative markers were evaluated by immunoblotting. Proteomic differential analysis was performed by Label Free nLC tandem mass spectrometry analysis by nanoACQUITY UPLC (Waters) coupled on-line to a LTQ Orbitrap XL (Thermo Electron, Bremen, Germany). Peptide identification was performed by Mascot (MatrixScience, London, UK) against Uniprot/Swissprot DataBase, and protein quantification by Progenesis LC-MS (Nonlinear Dynamics Ltd., Newcastle upon Tyne, UK). Proteins matching at least two unique peptides were considered for the analysis. Proteins with
Results
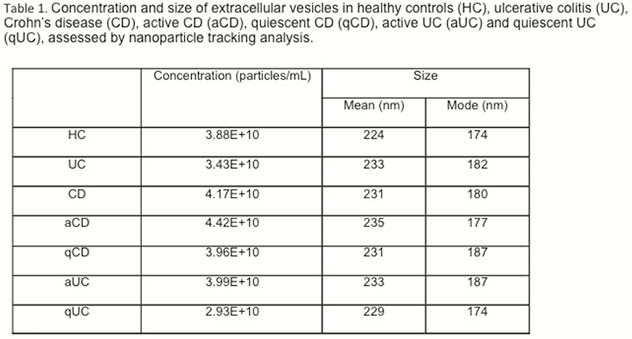
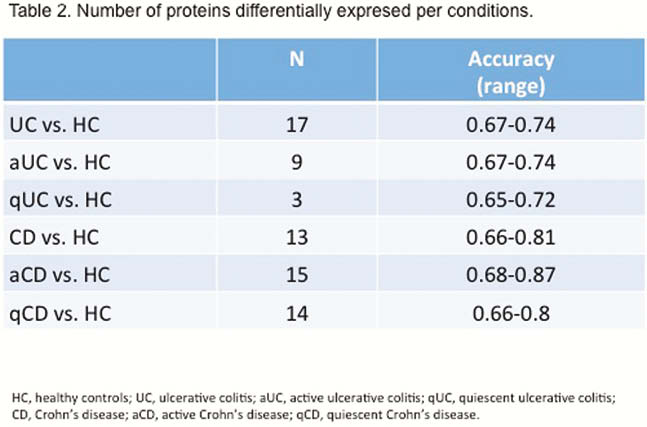
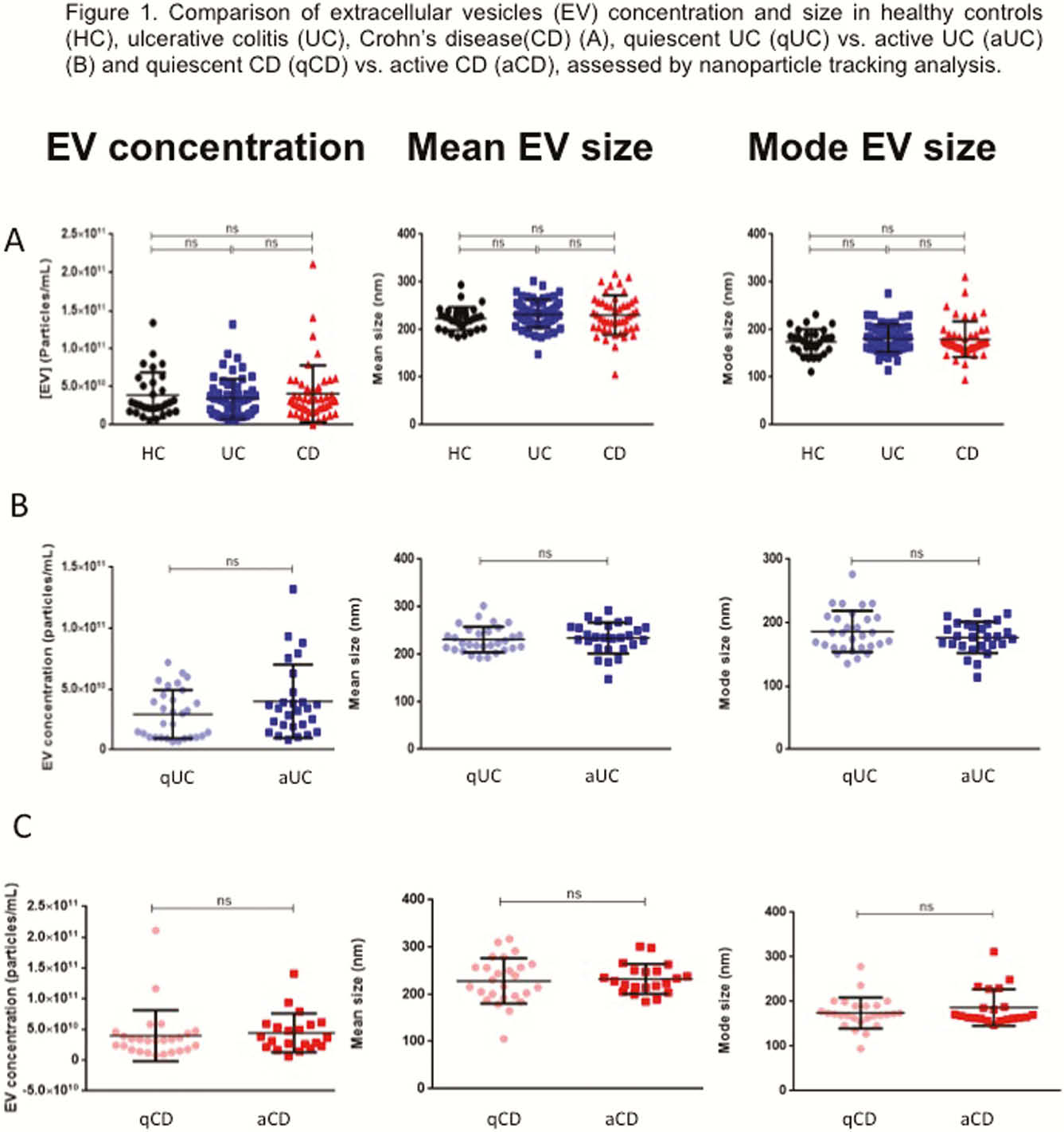
EVs size and concentration are shown in Table 1. There were not differences in EVs concentration and size between groups (Figure 1). A total of 324 proteins were identified, being some of them differentially expressed in different conditions (Table 2). Several proteins met the criteria to be proposed as accurate biomarkers: 17 in UC vs. HC, 9 in aUC vs. HC, 3 in qUC vs. HC, 13 in CD vs. HC, 16 in aCD vs. HC and 14 in qCD vs. HC. These proteins are involved in relevant processes in IBD pathogenesis such as angiogenesis, cell migration, immune response, acute phase response, inflammation and coagulation. Finally, a panel of seven proteins was chosen for validation in an independent cohort (Table 3).

Conclusion
Serum EVs of patients with IBD (CD and UC) carry differentially expressed proteins compared with HC and also depending on the presence of intestinal inflammation. These proteins are promising biomarkers of IBD that will be validated in an independent cohort.
M. Chaparro and M. Azkargorta have equally contributed as first authors. F. Elortza and J.P. Gisbert have equally contributed as senior authors.


