B. Verstockt1, S. Verstockt2, D. Alsaoud2, J. Sabino1, M. Ferrante1, S. Vermeire1
1IBD Leuven, 1Department of Gastroenterology and Hepatology, University Hospitals Leuven, Leuven, Belgium, 2KU Leuven, CHROMETA - Translational Research in Gastrointestinal Disorders, Leuven, Belgium
Background
The pan-Janus kinase (JAK) inhibitor tofacitinib (TFC) has recently been approved for and added to the treatment armamentarium of patients with moderate-to-severe ulcerative colitis (UC). With increasing choices and absence of predictive biomarkers generated from pivotal trials, real-world data are needed to stratify patients based on their molecular fingerprint in order to improve likelihood of response.
Methods
We obtained inflamed colonic biopsies from 52 consecutive patients initiating biological therapy (anti-TNF [n = 16], vedolizumab [VDZ, n = 20]) or TFC [n = 16] for active UC (Mayo endoscopic sub-score ≥2). Treatment choices were made in agreement between patient and physician, but all included patients were naïve for the mode of action initiated. All patients were treated with standard dosage according to the label. The response was defined as a Mayo endoscopic sub-score ≤1 and assessed by weeks 8–14. RNA was extracted and single-end RNA sequencing performed using Illumina HiSeq4000. Sequencing data were analysed through differential gene expression (DESeq2) and an unbiased network biology approach (Weighted Gene Coexpression Network Analysis).
Results
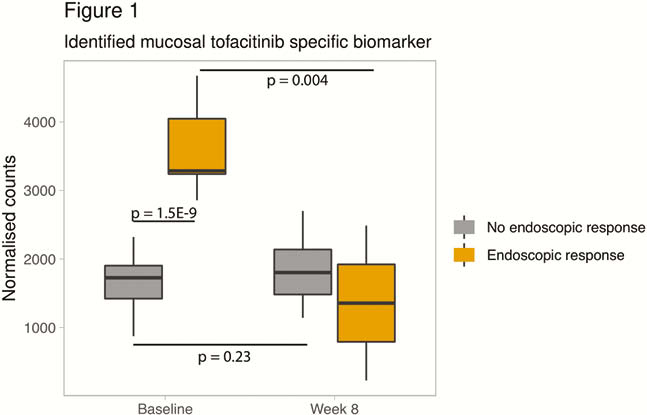
Response was observed in 5 (31.2%) anti-TNF, 13 (65.0%) VDZ, and 5 (31.2%) TFC-treated patients. Previously reported markers for anti-TNF response, including oncostatin M (p = 0.62), TREM1 (p = 0.70) and IL13RA2 (p = 1.0) could not differentiate TFC responders from non-responders. Similarly, the 4-gene vedolizumab signature [MAATS1, PIWIL1, RGS13 and DCHS2] could not discriminate TFC responders from non-responders (p = 0.13). No baseline differences in JAK/STAT-signalling could be identified either. Hence, we performed an unbiased network analysis of all TFC samples which identified 1 cluster of 65 genes, significantly correlating with response (p = 0.006). The hub gene within this network turned out to be the most differentially expressed gene (p = 1.5E−9, fold change [FC] 2.3), with a predictive accuracy for response of 100% (p < 0.001). In contrast, this gene could not predict anti-TNF or vedolizumab induced response (p = 0.13; p = 0.10, respectively). Of interest, baseline expression of the identified marker did not correlate with C-reactive protein, faecal calprotectin, Mayo endoscopic sub-score and other inflammatory markers including IL-6, IL-1B or epithelial markers. In TFC responders, the identified biomarker was significantly reduced by week 8, as compared with baseline (fold change [FC] -3.1, p = 0.0004), but not in non-responders (FC 1.2, p = 0.2) (Figure 1).
Conclusion
We identified a TFC-specific biomarker unrelated to disease severity, increasing the potency of a robust predictive marker based on its underlying mode-of-action. Validation in independent larger datasets is warranted.



