P696 Genetic predisposition to infliximab immunogenicity in patients with immune-mediated inflammatory diseases – secondary analyses from a randomised clinical trial
Bjørlykke, K.(1,2);Brun, M.K.(2,3);Viken, M.K.(4,5);Stenvik, G.E.(3);Klaasen, R.A.(6);Gehin, J.E.(2,6);Warren, D.J.(6);Sexton, J.(3);Haavardsholm, E.A.(2,3);Jahnsen, J.(1,2);Lie, B.A.(2,4,5);Goll, G.L.(3);Bolstad, N.(6);Syversen, S.W.(3);Jørgensen, K.K.(1);
(1)Akershus University Hospital, Department of Gastroenterology, Lørenskog, Norway;(2)University of Oslo, Institute of Clinical Medicine, Oslo, Norway;(3)Diakonhjemmet Hospital, Division of Rheumatology and Research, Oslo, Norway;(4)Oslo University Hospital, Department of Medical Genetics, Oslo, Norway;(5)Oslo University Hospital, Department of Immunology, Oslo, Norway;(6)Oslo University Hospital, Department of Medical Biochemistry, Oslo, Norway;
Background
Immunogenicity is a leading cause of treatment failure to TNF inhibitors. Genetic variations in HLA class II genes have been suggested to predispose to anti-drug antibody formation, but studies using high-resolution HLA typing as well as characterisation of biologically relevant haplotypes are currently lacking. The aim of this study was to assess associations between HLA loci and immunogenicity to infliximab.
Methods
Patients with immune-mediated inflammatory diseases (IMID) treated with infliximab (N=612) (120 rheumatoid arthritis, 181 spondyloarthritis, 72 psoriatic arthritis, 114 ulcerative colitis, 80 Crohn disease, and 45 psoriasis) were included in the randomised, Norwegian Drug Monitoring (NOR-DRUM) trial. Neutralising anti-drug antibodies were detected with an automated fluorescence assay. Next generation HLA sequencing at G group resolution were performed and trimmed to 2nd field. Haplotype analyses including the most significant HLA loci were performed, and epitope predictions for the infliximab light- and heavy-chain were assessed.
Results
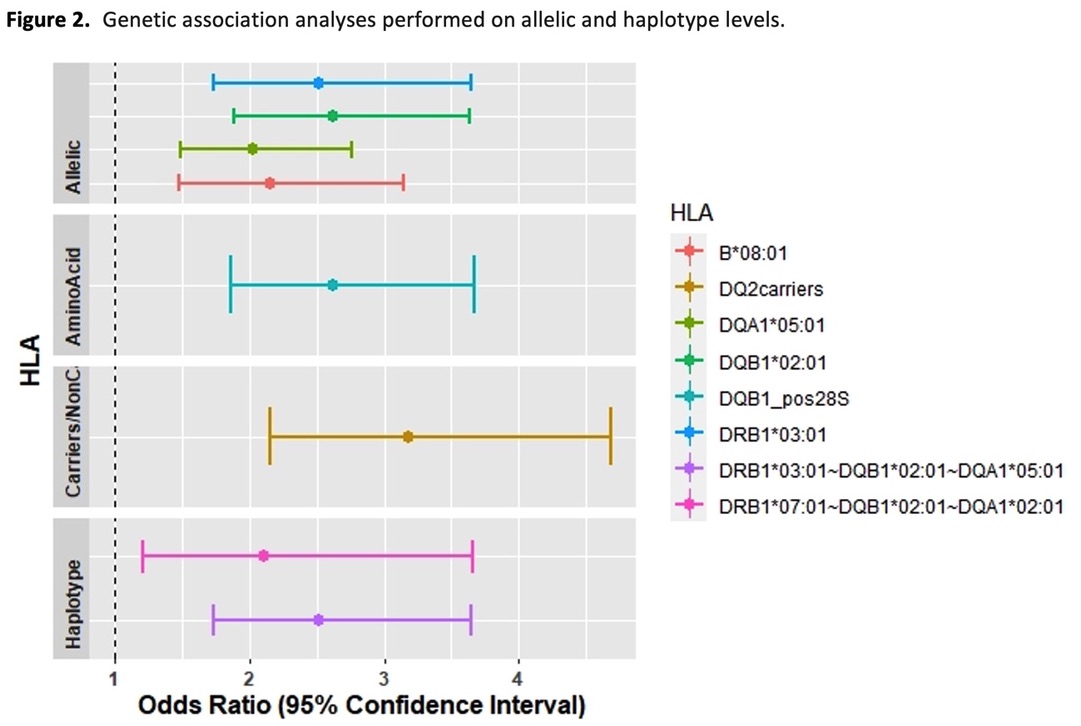
Anti-drug antibodies to infliximab were detected in 147 (24 %) of patients. The most significant associations with immunogenicity were with the genes HLA-DQB1 (P=1.4x10-6), -DRB1 (P= 2.7x10-5), -DQA1 (P=0.00017) and -B (P=0.0009). Conditional analyses indicated that HLA-DQB1 represents the primary association(Table 1). Haplotype analyses showed that there was significantly increased risk of immnogenicity in patients carrying the HLA-DQ2 haplotypes DQB1*02:01~DQA1*05:01 (P=1.1x10-6) and DQB1*02:02~DQA1*02:01 (P=0.008), while one protective haplotype DQB1*05:01~DQA1*01:01 (P=0.0005) was identified (Figure 1a-c). Presence of HLA-DQ2 more than doubled the risk of anti-drug antibody formation (HR 2.54, 95%CI 1.82-3.56) (Figure 1d), in a model including the covariates age, sex, diagnosis, and concomitant immunosuppressive therapy. Association tests of each level of HLA analyses showed that HLA-DQ2 represents the strongest association(Figure 2). Epitope predictions revealed that the HLA-DQ2 haplotypes DQB1*02:01-DQA1*05:01 and DQB1*02:02-DQA1*02:01 share nine strong binder peptides, originating from the infliximab light chain, encompassing the amino acid core-peptide sequences INTVESEDI and VYACEVTHQ.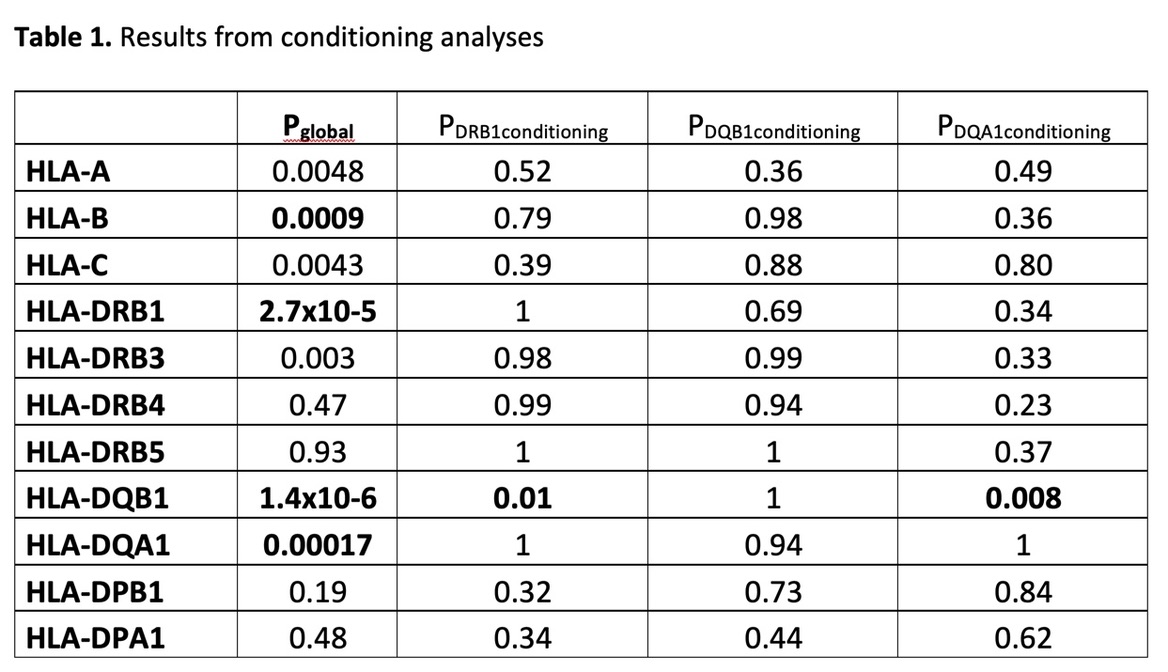

Conclusion
Using high-resolution HLA sequencing, we here demonstrate a genetic association between HLA-DQ2 and infliximab immunogenicity in IMID patients. The presence of either HLA-DQ2 risk haplotypes more than doubled the risk of anti-drug antibody formation across all diagnoses. Testing for HLA-DQ2 could be of clinical value in identifying patients at risk for immunogenicity and facilitate treatment decisions. Whether these findings apply to other TNF inhibitors should be further explored.


