P825 The impact of a SNP-dose–response effect on course of inflammatory bowel disease: a numerical analysis of SNP distribution within the Swiss IBD Cohort Study
L. Biedermann1, L. Denoth1, J.B. Rossel2, M. Butter1, S. Vavricka3, B. Misselwitz4, T. Greuter1, P. Schreiner1, I. Cleynen5, G. Rogler1, M. Scharl1
1University Hospital Zurich, Gastroenterology and Hepatology, Zurich, Switzerland, 2University of Lausanne, Center for Primary Care and Public Health Unisanté, Lausanne, Switzerland, 3Private Practice, Center for Gastroenterology and Hepatology, Zurich, Switzerland, 4Inselspital Bern, Gastroenterology and Hepatology, Bern, Switzerland, 5KU Leuven- Leuven- Belgium, Department of Human Genetics, Leuven, Belgium
Background
Several large genome wide association studies have identified an increasing number of IBD risk loci with currently more than 240 established loci and generated robust data on disease association. However, knowledge remains limited on distinctive associations to specific clinical characteristics. The Swiss IBD Cohort Study (SIBDCS) represents a large prospective cohort study with clinically and phenotypically sound data collection since 2006 including genotyping. In this study, we aimed to determine individual associations of known risk loci with clinical features of patients in the SIBDCS, as well as their combined effect on clinical outcomes.
Methods
Based on 158 analysed SNPs, we investigated the numerical distribution of risk alleles and determined an individual SNP risk score (defined as percentage of risk alleles; i.e. number of risk alleles divided by twice the number of given SNPs multiplied with 100). We then performed linear regression modelling to investigate, whether relevant clinical disease characteristics associate with this SNP risk score. Further, for each given clinical outcome, a model was run with all SNPs as potential predictors, and the number of significant associations per SNP counted.
Results
In a total of 2304 genotyped patients, we observed a median number of risk alleles of 167, 168 and 167 for IBD overall, CD and UC/IC, respectively with a narrow inter quartile range [11 (q25 = 162, q75 = 173) for IBD and CD; 12 (q25 = 161, q75 = 173) for UC/IC]. A higher SNP risk score was significantly associated with any complications (defined as a composite of any or more of colorectal cancer, colon dysplasia, intestinal lymphoma, osteopenia/-porosis, anaemia, deep venous thrombosis, pulmonary embolism, nephro- or cholelithiasis, malabsorption syndrome, massive haemorrhage, perforation/peritonitis, pouchitis), stenosis in CD patients; pancolitis, conversion to CD, female sex in UC patients; higher clinical disease activity in both CD & UC.
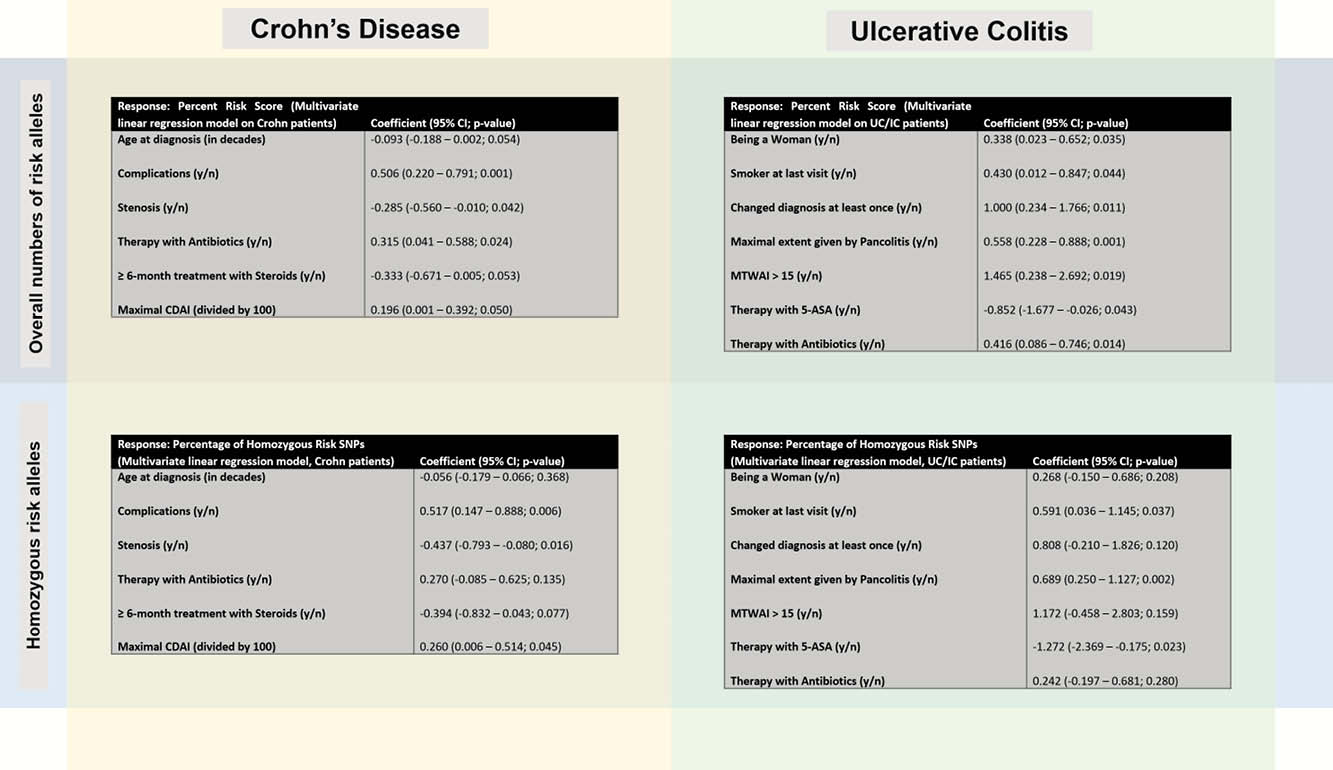
Regarding individual SNPs, we identified substantial differences in terms of the frequency of associations to disease-related outcomes with up to 11 for rs4899554 in CD and 7 for rs9557195 in UC, respectively.
Conclusion
In our large population of IBD patients there is high per patient frequency of hetero- and homozygous SNP risk alleles. The association of higher SNP risk score with several disease-related outcomes indicates a potential interplay of per patient given SNP risk alleles.


