P904 Genetic Variants in IBD Chilean Patients are related to clinical outcomes.
Pérez, T.(1,2)*;Bustamante, M.L.(3);Travisany, D.(4);Magne, F.(3);Hernandez-Rocha, C.(1);Alvarez-Lobos, M.(1);Orellana, M.(5);Ascui, G.(6);Azocar, L.(1);Espino, A.(1);Aguilar, N.(1);Baez, P.(7);Zazueta, A.(3);Estela, R.(2);Silva, V.(2);Escobar, S.(2);De la Vega, A.(2);Arriagada, E.(2);Miquel, J.F.(1);Alvares, D.(8);
(1)Pontificia Universidad Católica de Chile, Department of Gastroenterology, Santiago, Chile;(2)Hospital San Borja Arriarán, Department of Gastroenterology, Santiago, Chile;(3)University of Chile, Faculty of Medicine- ICBM, Santiago, Chile;(4)Universidad de las Americas, NIDS- Núcleo de Investigación en Data Science- Facultad de Ingeniería y Negocios-, Santiago, Chile;(5)University of Chile, Department of Computer Science- Faculty of Physical Sciences and Mathematics., Santiago, Chile;(6)La Jolla Institute for Immunology, Immunology, San Diego, Chile;(7)University of Chile, Center for Mathematical Modeling, Santiago, Chile;(8)University of Cambridge, MRC Biostatistics Unit, Cambridge, United Kingdom;
Background
IBD has emerged as a worldwide disease with an increased incidence in newly industrialized countries such as Chile. This population has been underrepresented in genome-wide association studies (GWAS) because IBD genetics studies have focused mainly on North America and European populations. In addition, specific IBD risk alleles have been related to different races and ethnicities. Aim: To investigate the association of IBD risk variants reported in previous GWAS studies with clinical outcomes in our Chilean patients.
Methods
192 Chilean individuals with IBD (145 UC and 47 CD) were genotyped using Illumina GSA Arrays. From IBD GWAS (Jostin et al. and Liu et al.), we selected gene variants with pGWAS value < 5x10-8 and looked for them in our Chilean IBD group. Then, we built a Chilean dataset (clinical-genotype information). Using this dataset, we performed a Spearman correlation matrix to correlate clinical outcomes with IBD variants. Further, we built regression models to predict the clinical outcomes using the variants obtained from the correlation matrix (p <0.05). Then, we selected the best models using significance testing (P values) or likelihood-based information criterion, such as the Akaike Information Criterion (AIC) and plotted the models using a Receiver Operating Characteristic Curve (ROC). Finally, to evaluate the association among variants in each model, we perform a Gene Ontology biological process enrichment analysis using PANTHER (Fisher, FDR).
Results
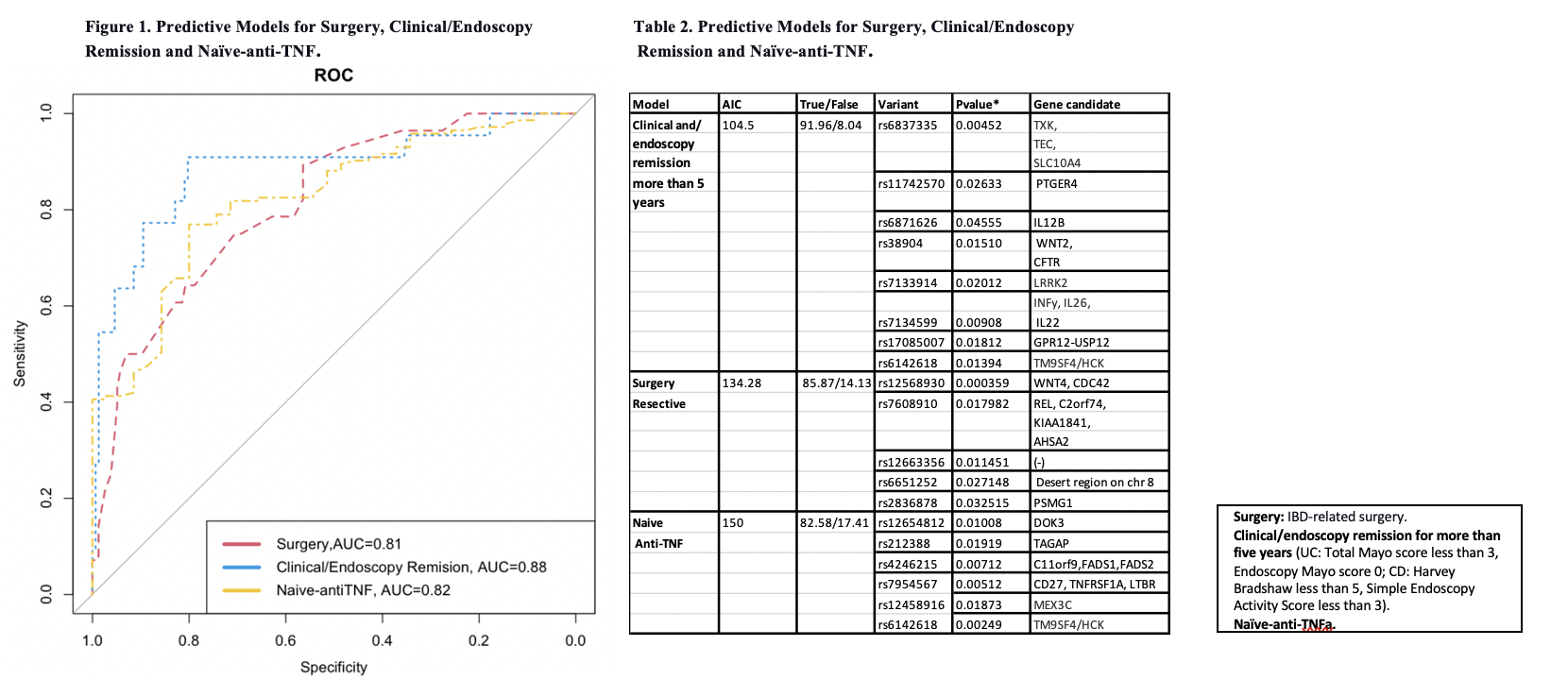
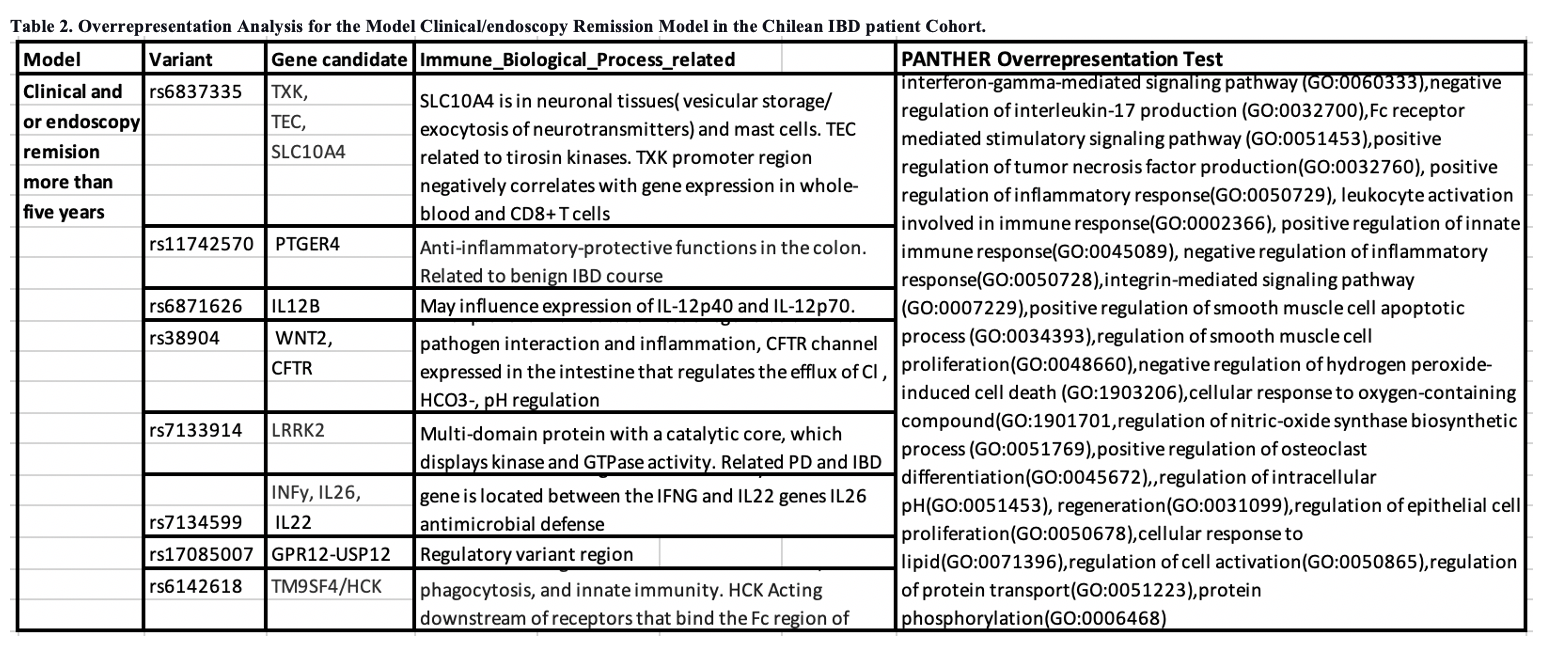
As shown in Figure 1 and Table 1, the best predictive regression models (more than 80%) for the clinical outcomes were surgery, Clinical/Endoscopy remission for more than five years, and Naïve anti-TNF. Association with genetic variants was observed significantly (p<0.05) in the enrichment analysis for the model Clinical/endoscopy remission (Table 2).
Conclusion
Conclusion. Candidates' genes related to clinical outcomes in our Chilean IBD cohort were related to epithelial, innate, and adaptative immune responses and host-microbial interactions. Future research is needed to validate these findings.
- Posted in: Poster Presentations: Genetics 2023


