6th EpiCom Workshop: Overview of prognostic factors and outcomes in IBD
Ravi Misra, EpiCom Member
 Ravi Misra Ravi Misra© Ravi Misra |
The Epidemiological Committee (EpiCom) organised the 6th EpiCom Workshop at the ECCO’22 Virtual Congress. The delegates were welcomed by EpiCom chair, Naila Arebi.
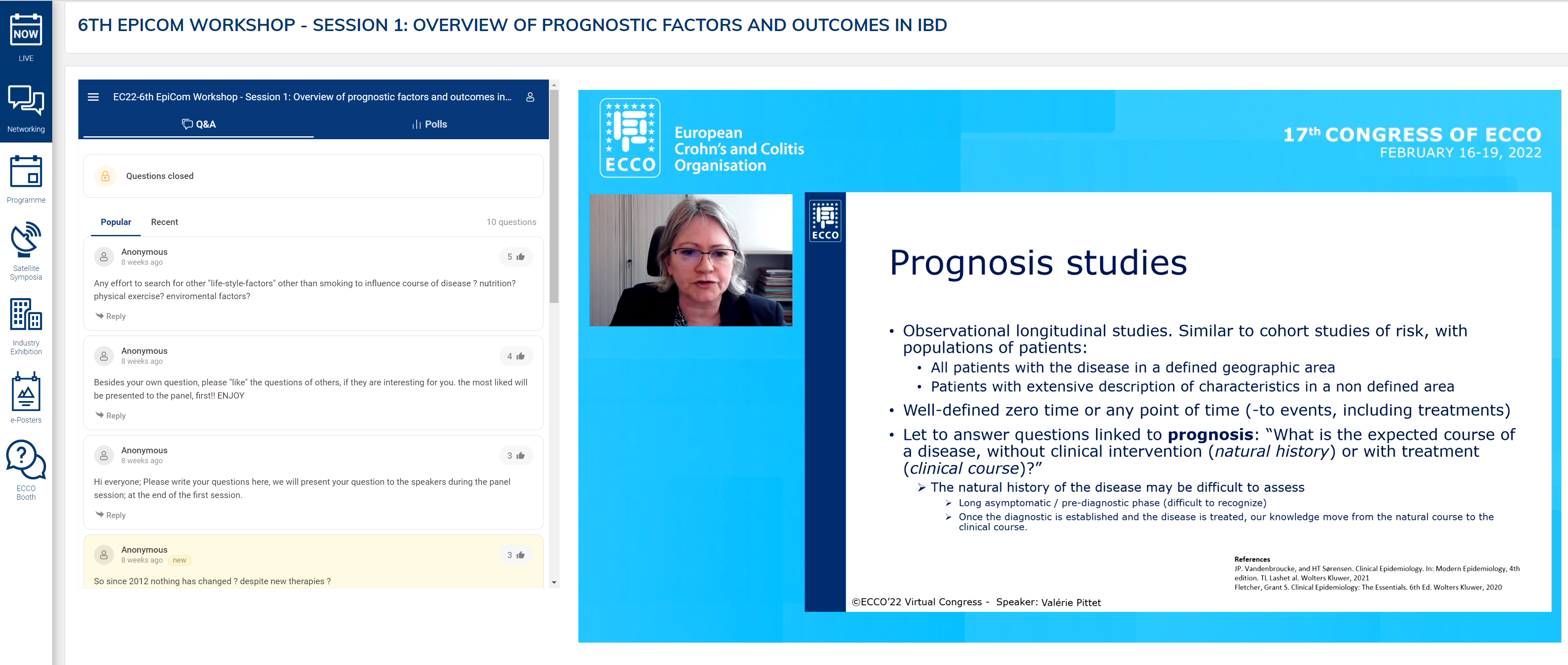
The opening presentation was a tandem talk by Naila Arebi and Valérie Pittet on core outcomes relevant to prognostic factors. The two different types of prognostic study, natural history and clinical course, were described, with identification of their strengths and weaknesses. Emphasis was placed on the importance of these studies in paving the way for personalised medicine by identifying high-risk groups and tailoring treatment accordingly. However, a significant limitation is the heterogeneity of outcomes, which limits comparison. Indeed, over 200 different outcomes have been described for Crohn’s Disease (CD) alone.
These findings demonstrate the need for a core outcome set (COS). A COS is defined as an agreed set of outcomes that should be measured and reported in all studies in a clinical area. Initial examples were OMERACT in rheumatoid arthritis. The Core Outcome Measurement in Effectiveness Trial (COMET) was launched in 2010 to make it easier to compare, contrast and combine studies. Using the COMET methodology, a COS for fistulising CD was created. The question is how this methodology can be applied to real-world data.
Naila Arebi discussed the barriers to developing a COS for prognosis, namely the large number of outcomes. Our practice has changed over time, with more focus on endoscopic, histological and patient-reported outcomes than before. Therefore, future observational studies should reflect the breadth of these outcomes. The inconsistency of studies in predicting risk of surgery in CD according to disease location illustrates the need to pool data, the feasibility of which depends on the collection of similar outcomes across studies. Unreliability in terminology limits the ability to compare studies. The differences between predictive, risk and prognostic factors were described. Poor prognostic factors for Ulcerative Colitis (UC) are young age at diagnosis and disease severity at diagnosis. The emphasis on biomarkers and histology helps to guide prognosis in Acute Severe Colitis. A recent study described 31 prognostic factors for advanced colorectal cancer in UC – but only one had strong evidence. With regard to CD, prognostic factors are applied to determine whether a top-down or step-up approach is required. In summary, although prognostic markers are clearly relevant to clinical practice, they are not weighted according to strength of evidence and in future the focus should be on molecular markers to predict disease course.
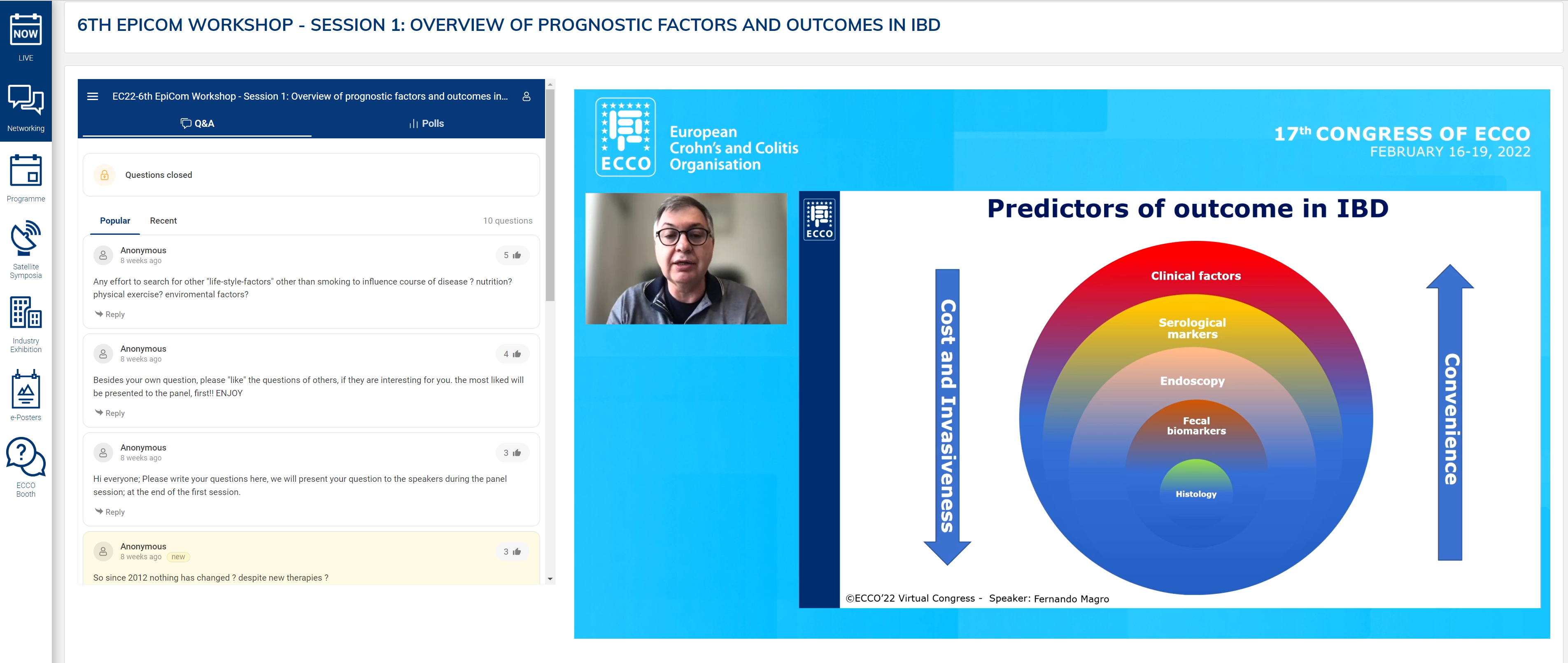
Fernando Magro presented an in-depth analysis of disease risk factors as prognostic tools. In summarising the prevailing data on prognostic studies, the most important difference was length of follow-up. Although several types of genetic and microbiological markers exist, current clinical practice is dependent on clinical risk factors for prognostication.
Fernando Magro described a risk matrix of disability in CD using only clinical markers. In patients with perianal disease, B3 with surgery within 6 months, immunosuppression and age less than 40 years, the likelihood of disabling disease is 91%. The same factors were associated with a 51% risk of re-operation. A systematic review of 42 paediatric studies revealed older age at presentation, small bowel and perianal involvement and ethnicity to be risk factors for stricturing and penetrating disease. Serological markers can be used to obtain risk for B2 and B3 disease behaviour.
Protein markers can also predict short-term flare after withdrawal of infliximab, as evidenced in the STORI trial. In combination, clinical and serological markers can offer statistically significant prognostic potential. Clinical risk factors in UC were described. Female gender is protective, as is smoking. Factors associated with a more severe phenotype were extensive disease and requirement for hospitalisation or steroids.
In summary, precision medicine with genetic or serological tests may allow early adjustment of treatment to improve prognosis. Baynesian networks may help to aggregate the information where statistical independence is not hidden.
The following tandem talk by Sophie Restellini and Julien Kirchgesner was entitled “Prognostic factors for therapy (Drug and Surgery)”.
The benefit of prognostication is that it enables us to individualise and make the best therapeutic decision at the correct time. Biomarkers can assist in the decision-making process but currently few are employed in clinical practice. Barriers to identifying a good biomarker were described: the need to take samples at diagnosis for prediction, large sample size, longitudinal sample collection, prospective follow-up and validation of the biomarker, ideally in a separate cohort. However, biomarkers for therapeutic efficacy have been discovered; for infliximab, HLA variants can predict risk of immunogenicity and single gene transcriptomic factors can distinguish non-responders. Growing interest in integrating multiple data sources will help us find better biomarkers.
Julien Kirchgesner focused on the predictive markers for relapse after surgery. Clinical risk factors of poor surgical outcome are steroid use, malnutrition and penetrating disease. The REMIND study showed how specific bacterial taxa were associated with increased risk of post-endoscopic recurrence. This was validated in an external dataset. Postulated surgical techniques such as mesenteric resection and use of Kono-S anastomosis suture have been described as possible abrogating factors for relapse.
Regarding the development of biomarkers, the importance of internal and then external validation was described. An example of an appropriate performance measure of predictive biomarker models is the AUC curve to discriminate between the two outcomes. Calibration is the extent to which the predictive probability matches the actual probability. The model performance will be determined by the choice of threshold and should be chosen according to clinical context. Further analysis in the form of decision curve analysis shows how differences in threshold may affect decision making. Finally, the effect of biomarkers and their benefit for real-life clinical decision-making needs to be assessed to ensure an incremental benefit to clinical outcomes.
In the second session the implementation of prognostic factors to manage and modify factors was illustrated with case-based discussion. Sophie Restilleni and Julien Kirchgesner presented the case of a patient with UC. The patient had several risk factors for severe disease: young age at diagnosis, extensive colitis, obesity (associated with reduced response to anti-TNF) and male sex. Biomarkers for predicting a worse outcome were highlighted: haemoglobin, C-reactive protein (CRP) and albumin. The risk of venous thromboembolism and the current lack of accurate predictors were discussed. C. difficile is noted to be a poor independent predictor. Subsequent endoscopy for disease assessment in this patient showed progression of disease with UCEIS 6. UCEIS is more reliable, with low inter-individual variation compared to Mayo scoring. In terms of assessing response, calprotectin is more sensitive than CRP.
This clinical case required biologic treatment with infliximab, and the issue of how to communicate risk to the patient was tackled. In discussions with the patient it is better to refer to absolute risk rather than to relative risk in comparison with the general population In discussions with the patient it is better to refer to absolute risk rather than to relative risk in comparison with the general population The use of pictograms to illustrate lymphoma risk with azathioprine is particularly effective.
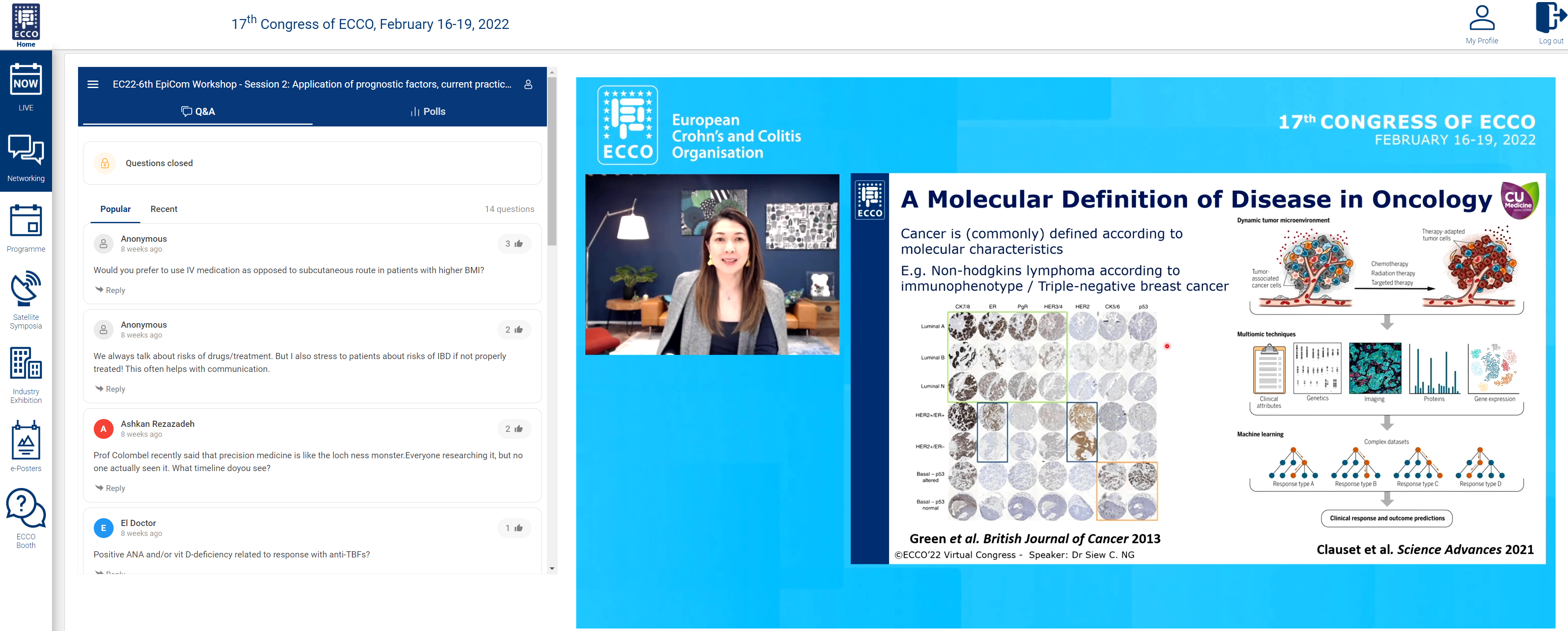
We were fortunate to end the session with a state of the art talk from Siew Ng on “Precision medicine to improve outcomes – dream or reality?”
Prediction of disease course through ‘-omic’ approaches was illustrated in a study of paediatric CD in 913 patients. Integration of clinical and serological data and transcriptomics showed upregulation of specific ileal genes, and bacterial species were associated with structuring and penetrating disease. Prediction of disease response was has been shown where overexpression of the cytokine oncostatin M was associated with anti-TNF resistance. Analysis of HLA-DQ alleles can be used to predict risk of immunogenicity to anti-TNF. This risk can be negated by use of an immunomodulator.
Tools to predict drug safety are already being used in clinical practice. TPMT and NUDT15 can predict myelosuppression in patients receiving azathioprine. Recent advances have helped to integrate the microbiome in IBD precision medicine. Defined patterns of microbial disturbances can predict relapsing CD. Metabolic function may predict response to certain drugs; for example, higher rates of butyrate synthesis are associated with higher responsiveness to anti-TNF.
There are significant challenges in identifying biomarkers. Biorepositories are required with large prospective clinical trials and there is a need to replicate these methods in external validation cohorts. Low cost, rapid availability of the results and high accuracy are needed for biomarkers to become a routine part of clinical care. However, multi-omic data integration studies have the potential to drive precision medicine in order to improve outcomes, and this potential could become a reality.
Browse through the gallery:
Pictures are subject to copyright © ECCO