P078 Proteomics and Lipidomics Analysis Revealed Alterations of Complement Activation and Lipid Metabolism in peripheral blood mononuclear cells of Inflammatory Bowel Disease Patients
Notararigo, S.(1);Bravo, S.(1);Martin-Pastor, M.(2);Baston-Rey, I.(3);Quiroga-Castiñeira, A.(3);Dominguez-Munoz, J.E.(3);Domínguez Medina, E.(1);Barreiro-de Acosta, M.(3);
(1)University Hospital Santiago De Compostela CHUS, Instituto de Investigación Sanitaria de Santiago IDIS, Santiago De Compostela, Spain;(2)CACTUS, Unidade de Resonancia Magnética, Santiago, Spain;(3)University Hospital Santiago De Compostela CHUS, Department of Gastroenterology- IBD Unit, Santiago De Compostela, Spain
Background
The lack of interaction between immune system cells and microbiota is considered a key driver of the pathogenesis of inflammatory bowel disease (IBD). The association of age-associated B cells (ABC) from antigen-experienced B cells and autoimmunity is now well established. The increased demands in lipid metabolism during immune response and cell activation upon autoimmune signals leads to lipid tag modifications of proteins. In IBD patients, we observed an impairment of B cell development, identifying a transitional B cell subset expressing CD38Hi, CD24Hi and CD19+. We hypothesize that metabolic alterations of proteins and lipids during immune cell activation might be a relevant mechanism of autoimmune disease.
Methods
An IBD cohort of patients in clinical remission under anti-TNF treatment was included in order to test metabolic and proteomic changes in peripheral blood mononuclear cells (PBMCs). Proteomic analysis was performed by Mass Spectrometry using a label free quantitative method (SWATH-MS analysis) and lead to identify a number of proteins underregulated (< 0.6) and overexpressed (> 1.5) fold change respect to control. Lipidomics of serum samples analyzed by proton Nuclear Magnetic Resonance spectroscopy (NMR)2 and multivariant statistics.
Results
Serum samples were obtained in Crohn disease (CD) n = 18, ulcerative colitis (UC) n = 9, before biological infusion and healthy controls (CTRL) n = 10. Proteins related with lipid metabolism like APOC2 for CD and APOB, APOA1 for UC, were differentially modulated. Significant differences in non-polar metabolites and lipids that phenotypically define CD and UC groups respect to CTRL were observed (Fig. 1)
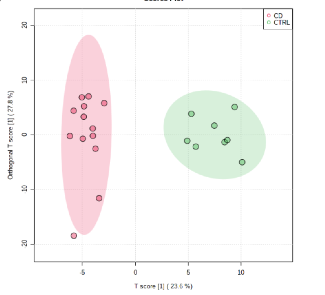
Fig. 1 Orthogonal-Partial Least Square-Discriminant Analysis (OPLS-DA) of the global NMR data of the serum samples of groups CD and CTRL.
Indeed, proteins associated with complement activation like Complement Factor 1 and Complement C1q subcomponent subunit B were differentially expressed in CD group. These results were confirmed by lipidomic analysis, in which palmitic (Fig. 2) as well as other candidate lipids were more abundant in IBD groups than CTRL. 
Fig2 Differences of palmitic acid concentration in CD and UC patients vs CTRL (Bonferroni test showed statistical difference between CTRL vs. CD and CTRL vs. UC with p = 0.02 and p = 0.004 respectively).
Conclusion
Our study detected high levels of proteins regulating lipid metabolism and palmitic in IBD patients. This observation can be interpreted as part of deleterious alterations of proteins through palmitoylation during B-cell activation and might be a relevant mechanism of disease. Further exploration of palmitate catabolism might shed light to the mechanisms that result in immunometabolic disorders.
- Posted in: Poster presentations: Basic Science 2021


